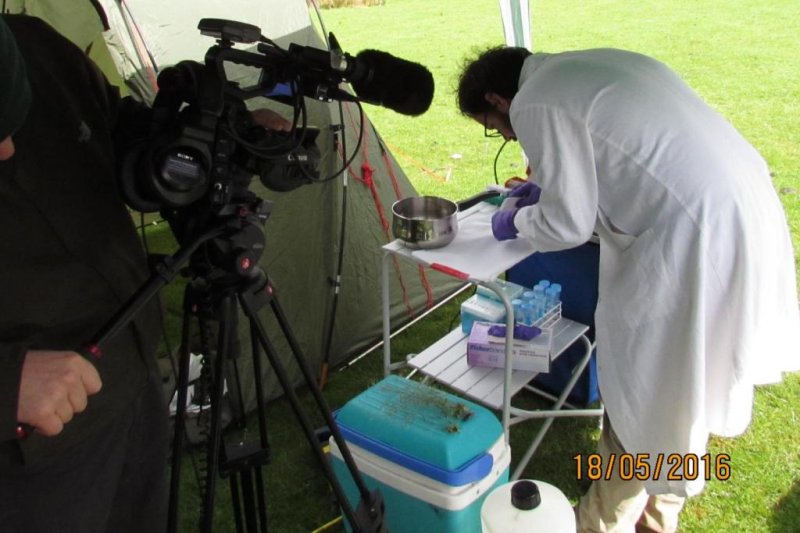
Alex Papadopolous, a scientist at Royal Botanic Gardens, Kew, sets up a mobile lab in the Welsh Mountains to test the handheld MinION sequencer in plant identification for the first time. Photo by Alex Papadopolous
Aug. 21 (UPI) -- Scientists at the Royal Botanic Gardens, Kew, have developed portable, real-time DNA sequencing that can be conducted on location, such as a mountainside, they report in a new study.
"This research proves that we can now rapidly read the DNA sequence of an organism to identify it with minimum equipment," Joe Parker, a scientist at Kew, said in a press release.
"Rapidly reading DNA anywhere, at will, should become a routine step in many research fields. Despite hundreds of years of taxonomic research, it is still not always easy to work out which species a plant belongs to just by looking at it. Few people could correctly identify all the species in their own gardens."
The study, published today in Scientific Reports, allowed scientists to sequence a whole genome on a Welsh mountainside to identify a plant species within hours.
For the past 40 years, DNA sequencing has changed the scientific world but is still a laboratory-bound practice, however, species identification is largely a field-based practice.
Researchers used the portable DNA sequencer known as the MinION from Oxford Nanopore Technologies to analyze plant species in Snowdonia National Park, the first time genomic sequencing has been performed in the field.
Using the MinION, the team was able to identify two innocuous white flowers, Arabidopsis thaliana and Arabidopsis lyrata ssp. petraea.
"Accurate species identification is essential for evolutionary and ecological research, in the fight against wildlife crime and for monitoring rare and threatened species," said Alexander Papadopulos, also a scientist at Kew.
"Identifying species correctly based on what they look like can be really tricky and needs expertise to be done well. This is especially true for plants when they aren't in flower or when they have been processed into a product. Our experiments show that by sequencing random pieces of the genome in the field it's possible to get very accurate species identification within a few hours of collecting a specimen."